Keywords: Biopharmaceuticals, DSC, proteins, macromolecules
MC174
Understanding the energetic processes that dictate the structure, stability, and affinity of proteins and other macromolecules has achieved new importance with the recognition that minute alterations in these properties can dramatically alter the function of the biopolymer, often with severe physiological consequences. The successful engineering of proteins with desired novel properties, and the rational design of drugs that selectively bind to a protein or nucleic acid, require that the thermodynamic effect of a chemical alteration or genetic mutation be predicted with considerable confidence.
Differential Scanning Calorimetry (DSC) measures the change in energy in a sample as the temperature is raised or lowered, and thus can determine absolute thermodynamic data for thermallyinduced transitions. All macromolecules will denature when heated, and many proteins denature when cooled. Ultra-sensitive calorimeters quickly quantify the thermodynamic parameters (and hence stability) of proteins and nucleic acids in their native state, following mutation, or when bound to a drug candidate. Calorimetric measurements are direct: the intrinsic thermal properties of the sample a remeasured without the need for extensive purification, chemical derivatization, immobilization or spectroscopic probes, and if the unfolding process is reversible, the technique is non-destructive. Depending on the complexity of the unfolding profile, only nanomoles of material may be required to provide a complete analysis of thermally-induced transitions. Importantly, DSC data provides fundamental thermodynamic insights into the stabilizing and binding interactions crucial for rational protein and drug design.
This introductory note provides an overview of the types of biophysical problems that can be addressed using DSC, and briefly describes how a typical experiment is conducted and how the data is interpreted.
Applications of DSC
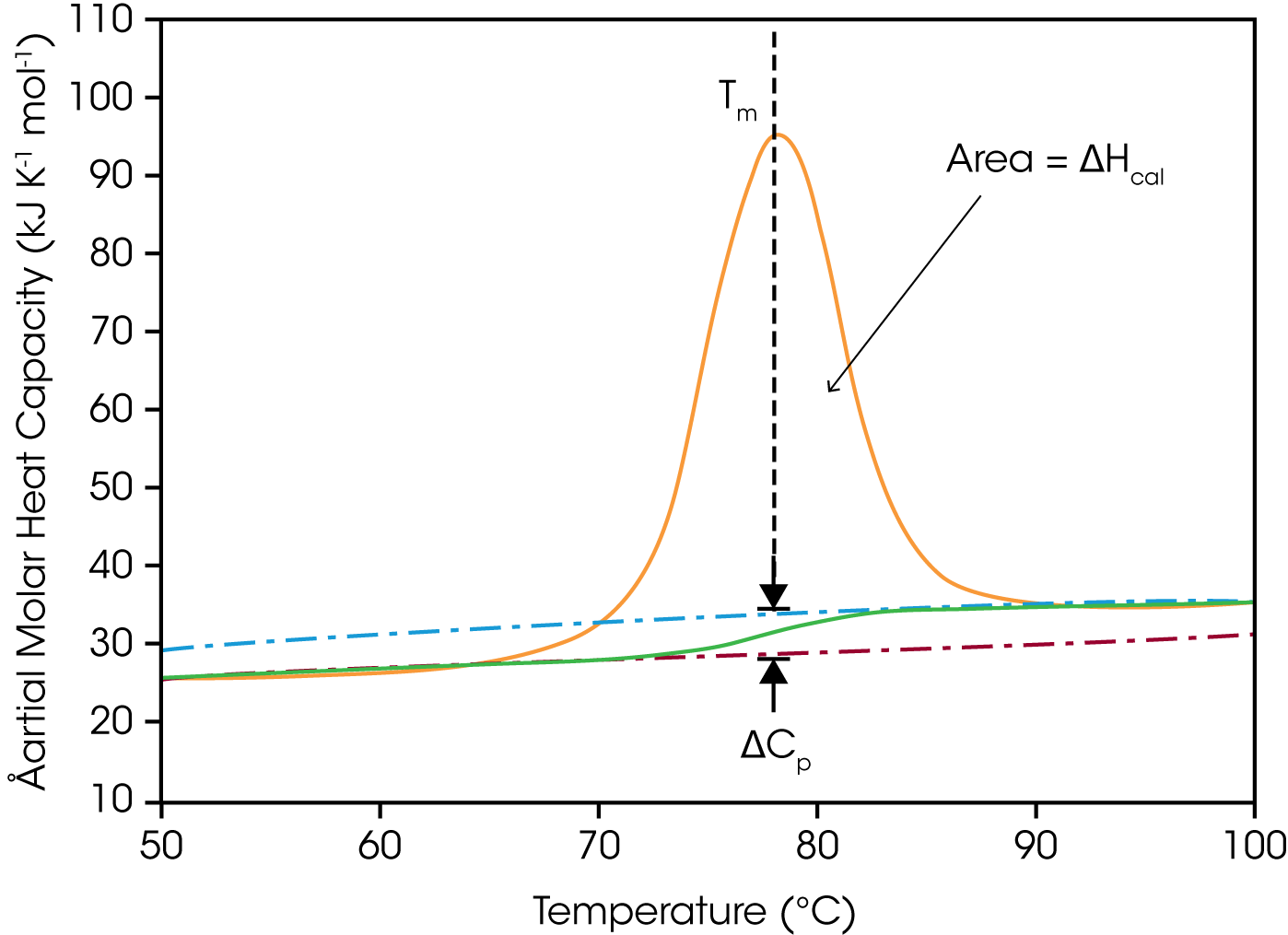
DSC is used in the life sciences to study the stability of macromolecules either alone in solution, or when bound to small ligands such as drugs or to large ligands such as other proteins or nucleic acids. Biological macromolecules in solution are constantly undergoing conformational fluctuations and are in equilibrium between the native, folded conformation and an ensemble of energetically similar unfolded conformations. Whether a macromolecule is folded or unfolded under a given set of conditions depends on the relative contributions of the enthalpic (ΔH) and entropic (ΔS) components of the system (i.e., ΔG= ΔH – TΔS). Unfolding occurs when TΔS increases sufficiently (for example, by the absorption of heat) to overcome stabilizing enthalpic interactions such as hydrogen bonds, hydrophobic interactions and electrostatic interactions, giving rise to an endothermic peak.
In a single thermal unfolding experiment, DSC can directly measure and allow calculation of all the thermodynamic parameters characterizing a biological molecule: the ΔHcal (the enthalpy as measured by calorimetry) due to thermal denaturation, the transition midpoint (Tm here half the macromolecules are folded and half are unfolded), the change in heat capacity (ΔCp) due to thermal unfolding, p the Gibbs free energy (ΔG) and the entropy (ΔS). DSC is thus, a powerful approach to studying:
- Protein Stability
- Protein Domain Structure
- Nucleic Acid Stability
- Biopolymer Conformation and Solvation
- Membrane Stability
- Stabilizing Effects of Ligand Binding
Design of a Typical DSC Experiment
The following example describes a general DSC protocol for studying protein stability, but a similar approach would be used with any biological molecule. A DSC instrument such as the Nano DSC contains two matched cells (a sample cell and a reference cell) that are heated at identical rates. A buffer baseline scan is first obtained to establish the linearity of the baseline (buffers with high temperature dependencies and thermally unstable additives such as azide should be avoided). The protein is then scanned: a thoroughly dialyzed, degassed protein solution of known concentration (for example, a 10 μM buffered solution) is loaded into the 0.3 ml sample cell, and the reference cell is filled with an equal amount of degassed dialysis buffer. User-defined variables (starting and ending temperatures, heating and cooling scan rate, number of scans, etc.) are input, then the instrument software takes control and automatically performs the entire experiment. The cells are pressurized at constant pressure (to prevent bubble formation during heating and allow determination of ΔCp) thermally equilibrated at the chosen starting temperature, then heated and cooled repeatedly as pre-specified by the user. Differences in heat uptake or production by the sample and reference cells are measured and compensated for by the instrument, thus maintaining both cells at the same temperature. The instrument provides a constant readout of the difference in heat absorption or release by the two cells, which corresponds to differences in the apparent heat capacities of the sample and reference. After cooling both cells to the starting temperature, the experiment can be repeated to determine if the biopolymer exhibits reversible folding/unfolding. Because of the extreme stability of buffer baselines obtained on Nano DSC’s subtraction of the buffer baseline corrects the data for the partial molar heat capacity of the solvent, allowing the partial molar heat capacity of the protein to be directly determined from the thermogram. The software package supplied with the DSC performs this subtraction, and also calculates the calorimetric enthalpy and entropy change of the protein. In addition, the data can be fit to two-state or multiple processes to determine the concentrations of folded and unfolded protein in the sample. Thus, a single automated DSC run requiring a fraction of a milligram of protein, about an hour per sample and limited operator attendance provides a complete thermodynamic analysis of the thermal stability of the protein.
Summary
In summary, DSC is a straightforward approach towards understanding the enthalpic and entropic changes that occur as a biopolymer is thermally denatured. Since the biopolymer can be free in solution (such as a protein or nucleic acid), associated with another molecule (as in a DNA/drug complex) or part of an assembly of molecules (such as lipids in a membrane), DSC can be used to study the stability of essentially any biological sample, including whole cells. Experiments are rapid, sample derivatization is not required, impurities are tolerated, and often only nanomoles of the target macromolecule are required.
Click here to download the printable version of this application note.

